Controlled Signaling and Transmitter Replenishment for MC with Functionalized Nanoparticles
Abstract
In this paper, we propose novel Transmitter (Tx) models for Molecular Communication (MC) systems based on functionalized Nanoparticles (NPs). Current Tx models often rely on simplifying assumptions for the molecule release and replenishment mechanisms. In contrast, we propose a Tx model where the signaling molecule release is controlled by a switchable membrane driven by an external trigger. Moreover, we propose a reloading mechanism, where signaling molecules are harvested based on an enzymatic reaction. Hence, no repeated injection of signaling molecules is required. For the proposed Tx model, we develop a general mathematical description in terms of a discrete-time transfer function model. Furthermore, we investigate two realizations of the proposed Tx model, i.e., an idealized Tx relying on simplifying assumptions, and a realistic Tx employing practical components for the reloading and release mechanisms. Finally, we numerically evaluate the proposed model and compare our results to stochastic Particle Based Simulations (PBSs).
Supplementary Material
Contents:
- Short summary of the proposed Transmitter (Tx) model including a description of the main features
- Short summary of the practical realization of the proposed Tx model
- Supplementary material for the Enzyme reaction and remarks on the implementation
1. System Model
As described in [1, Sec. 2], we propose a novel Transmitter (Tx) design for synthetic Molecular Communication (MC) systems employing functionalized Nanoparticles (NPs). The envisioned Tx comprises two main mechanisms namely
- a controlled release mechanism, which enables the release of signaling molecules controlled by an external stimulus (see Animation 1, left hand side),
- a reloading mechanism enabling the production of signaling molecules inside the Tx from molecules recruited from the surrounding environment (see Animation 1, right hand side).
Animation 1: Left hand side: Controlled release of signaling type B molecules (red) by controlled opening of the Tx membrane. Right hand side: Reloading of type A molecules (blue) from the surrounding environment and conversion into signaling type B molecules.
Both, the controlled release and the reloading are facilitated by the design and functionalization of the NP. The controlled release mechanism is realized by functionalization of the NP with a switchable membrane allowing for the control of its permeability by an external stimulus. The reloading mechanism is realized by enzymes encapsulated in the NP allowing for the conversion of molecules recruited from the surrounding environment, denoted as type A molecules, in signaling molecules, denoted as type B molecules. The switching of the NP membrane between the open and closed states is exploited for both the controlled release of type B molecules and the reloading of type A molecules, while the membrane is impermeable for enzymes in both states.
2. Practical Realization of the proposed Tx Model
In [1, Sec. 4] we describe a realization of the proposed Tx model based on practical components:
- The membrane switching for the controlled release is facilitated by polymersomes, the synthetic counterpart of liposomes, with a pH-driven permeability switch
- The reloading mechanism (signaling molecule production) is realized by the encapsulation of the enzyme Mandelate Racemase (MR) into the NP. MR performs a reversible one-substrate-reaction of (R)-Mandelate (type A molecules) to (S)-Mandelate (type B signaling molecule) The switching process of the NP is shown below in Animation 2, cf. [1, Fig. 2]
Animation 2: Reloading and release process of the practical Tx model. Process flow: Only type (R)man molecules (blue) and enzymes (green) are present in the environment; pH-decrease triggers the opening of the Tx membrane and (R)man molecules diffuse into the NP and are converted to (S)man signaling molecules (red); pH-increase triggers closing of the Tx membrane; pH-decrease triggers opening of the Tx membrane and the produced (S)man molecules are released while additional (R)man is harvested.
3. Supplementary Material - Enzyme Reaction
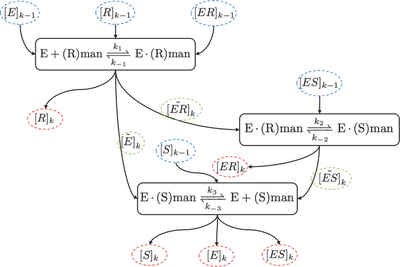
The enzyme Mandelate Racemase (MR) performs a reversible one-substrate-reaction of (R)-Mandelate (type A molecules) to (S)-Mandelate (type B signaling molecule), and the reaction equations are as follows [2,3] \begin{align} &\ce{E + (R)man <=>[$k_1$][$k_{-1}$] E*(R)man <=>[$k_2$][$k_{-2}$] E*(S)man <=>[$k_3$][$k_{-3}$]E + (S)man}, \, \mathrm{(12)} \end{align} where E denotes the enzyme MR, and E$\cdot$(R)man and E$\cdot$(S)man denote intermediate complexes of MR and (R)man and (S)man, respectively. The corresponding reaction rates are denoted by $k_1, \,k_2, \,k_3$ and $k_{-1}, \, k_{-2}, \, k_{-3}$, respectively.
Remarks on the Implementation
As described in [1, Sec. 4], (12) cannot be solved analytically. Instead we developed a numerical model that is evaluated in each discrete time step. The Python Code can be found on GitHub. We obtained a numerical model as follows:
Decomposition into three bidirectional reactions
First, we assume a pseudo steady state and derive a reaction rate equation for the three individual bidirectional reactions in [1, Eq. (12)] see, e.g., [4,5].
The individual bidirectional equations and the associated steady state reaction rate equations are as follows:
Reaction 1 The first bidirectional reaction in (12) follows as \begin{align*} &\ce{E + (R)man <=>[$k_1$][$k_{-1}$] E*(R)man} &&\mathrm{(12a)}. \end{align*} The associated reaction rate equation assuming a steady state follows as \begin{align*} &\frac{\partial [E]}{\partial t} = 0, &k_{-1}[ER] - k_1[E][R] = 0, &&\mathrm{(12b)} \end{align*} where [E], [R], and [ER] denote the concentration of MR, (R)man, and E$\cdot$(R)man, respectively.
Reaction 2 The second bidirectional reaction in (12) follows as \begin{align*} &\ce{E*(R)man <=>[$k_2$][$k_{-2}$] E*(S)man}. &&\mathrm{(12c)} \end{align*} The associated reaction rate equation assuming a steady state follows as \begin{align*} &\frac{\partial [ER]}{\partial t} = 0, &k_{-2}[ES] - k_2[ER] = 0, &&\mathrm{(12d)} \end{align*} where [ER] and [ES] denote the concentration E$\cdot$(R)man and E$\cdot$(S)man, respectively.
Reaction 3 The third bidirectional reaction in (12) follows as \begin{align*} &\ce{E*(S)man <=>[$k_3$][$k_{-3}$]E + (S)man}. &&\mathrm{(12e)} \end{align*} The associated reaction rate equation assuming a steady state follows as \begin{align*} &\frac{\partial [ES]}{\partial t} = 0, &k_{-3}[E][S] - k_3[ES] = 0, &&\mathrm{(12f)} \end{align*} where [E], [S], and [ES] denote the concentration of MR, (S)man and E$\cdot$(S)man, respectively.
For all three reaction rate equations (12b), (12d), (12f) a numerical model can be obtained straightforwardly, e.g., by numerical integration or by a solution in terms of an exponential approach. Further details on the exact numerical implementation of the individual bidirectional reactions can be found in the associated Python code.
Algorithm
Figure 1 shows the process of the developed algorithm to calculate the concentrations of all reaction partners in (12) by a sequential numerical evaluation of (12b), (12d) and (12f) in the current time step $k$ from the concentrations in the previous time step $k-1$, where we denote the concentrations in time step $k$ according to (12b), (12d) and (12f) as follows: \begin{align*} [E]_k, \, [R]_k, \, [S]_k, \, [ES]_k, \, [ER]_k. \end{align*}
References
[1] M. Schäfer, L. Brand, S. Lotter, A. Büyükoglu, F. Enzenhofer, W. Haselmayr, K. Castiglione, D. Appelhans and R. Schober, “Controlled Signaling and Transmitter Replenishment for MC with Functionalized Nanoparticles”, submitted to 9th ACM Int. Conf. Nanosc. Comp. Commun. , 2022, [online]: TODO
[2] M. St. Maurice and S. L. Bearne. 2002. “Kinetics and Thermodynamics ofMandelate Racemase Catalysis”. Biochem. 41, 12 (2002), 4048-4058.
[3] Ariun Narmandakh and Stephen L. Bearne. 2010. “Purification of Recombinant Mandelate Racemase: Improved Catalytic Activity”. Protein Expression and Purification, 69, 1 (2010), 39-46. https://doi.org/10.1016/j.pep.2009.06.022
[4] D. J. Higham. 2008. “Modeling and Simulating Chemical Reactions”. SIAMRev. 50, 2 (2008), 347-368. https://doi.org/10.1137/060666457
[5] L. A. Segel and M. Slemrod. 1989. “The Quasi-Steady-State Assumption: ACase Study in Perturbation”. SIAMRev. 31, 3 (1989), 446-477.